Statistical Learning Notes | Decision Tree 2 - Bagging & Random Forrest & Boosting
1.Bagging
– Out-of-Bag (OOB) Error Estimation
– Variable Importance Measures
2.Random Forest
– Decorrelating the Trees
3.boosting
– Algorithm
– Three Tuning Parameters
These three ensemble methods use trees as building blocks to construct more powerful prediction models
1. Bagging
Bootstrap is used when it is hard or even impossible to directly compute the Standard Deviation.
Bootstrap Aggregation (or bagging) can reduce the high-variance of decision trees
- high variance may result in fitting quite different trees on different part of same data
A set of ${X_1,…,X_n}\ $ with variance $\sigma^2$
Averaging a set of observations $\bar{X}\ $ reduces variance to $\sigma^2/n \ $
To reduce the variance of a model and improve the prediction accuracy
- Using B separate training sets from the population (hard since data may not enough)
- Fit separate model $\hat{f}^1(x),…,\hat{f}^B(x)$
- Average them to obtain a low-variance model:
$\hat{f}{avg} (x)= \frac{1}{B} \sum{b=1}^B \hat{f}^b(x)$
Bagging(Bootstrap) takes repeated samples from single trainging data set
- Generate B different bootstrapped training data sets
- then train the model on $b^{th}$ bootstrapped training set to get $\hat{f}^{*b}(x)$
- Average all the predictions:
$$\hat{f}{bag}(x)=\frac{1}{B}\sum{b=1}^B\hat{f}^{*b}(x)$$
Bagging for Regression Tree:
Construct B regression trees using B bootstrapped training sets, these trees are grown deep and unpruned
- each individual tree has high variance and low bias
- Average these B trees to reduce the variance and improve accuracy
Bagging for Classification Tree:
Record the class predicted by each of the B trees, and take a majority vote:
- The overall prediction is the most commonly occuring class among the B predictions.
Heart Disease Example:
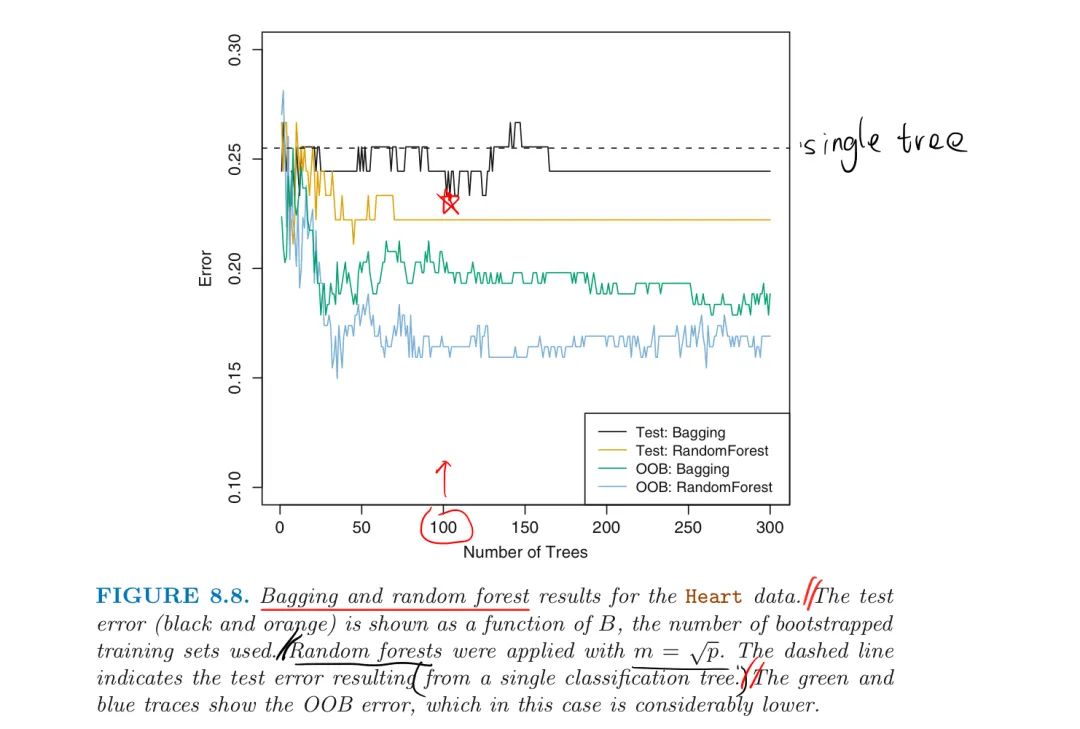
The number of trees B is not a critical parameter with bagging, large B will not lead to overfitting.
Out-of-Bag (OOB) Error Estimation
On average, each bagged tree makes use of around two-thirds of the observations,
- the remaining one-third of the observations not used to fit a given bagged tree are referred to as the out-of-bag (OOB) observations
For the $i,^{th}$ observation, there will be around B/3 predictions to average or take a majority vote.
The resulting OOB error is a valid estimate of the test error for the bagged model
- since the response for each observation is predicted using ONLY the trees that were NOT fit using that obseration
OOB replaces CV:
The OOB approach for estimating the Test Error is better when performing bagging on large data sets for which CV would be computationally onerous
- as B sufficiently large, OOB error is virtually equivalent to LOOCV error
Variable Importance Measures
Bagging improves accuracy at the expense of interpretability
- not clear which variables are important
Still can obtain an overall summary of the importance of each predictor by
- Record the total amount that the RSS is decreased due to splits over given predictor for bagging regression trees
- Add up the total amount that the Gini Index is decreased due to splits over given predictor for bagging classification trees
- Average over all B trees and a large value indicates an important predictor
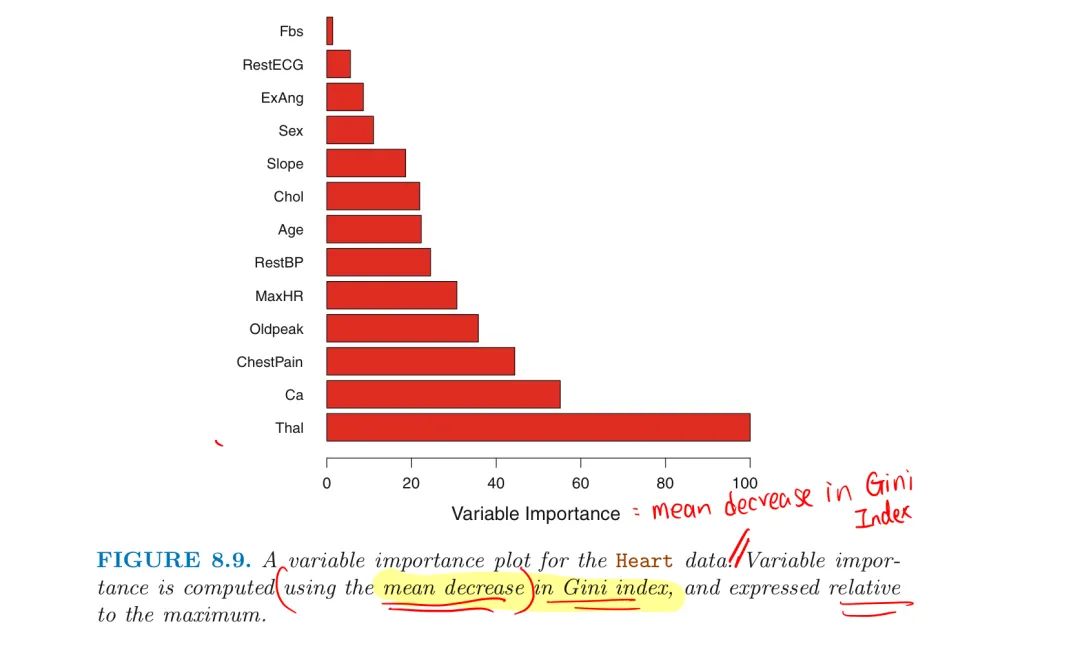
2. Random Forest
Random Forests provide an improvement over bagged trees by a way of a small tweak that decorrlates the trees.
As in bagging, RF builds a number of trees on bootstrapped training samples,
- a random sample of m predictors is chosen as split candidates from all p predictors
- a fresh resample of m predictors at each split and use ONE of those m predictors
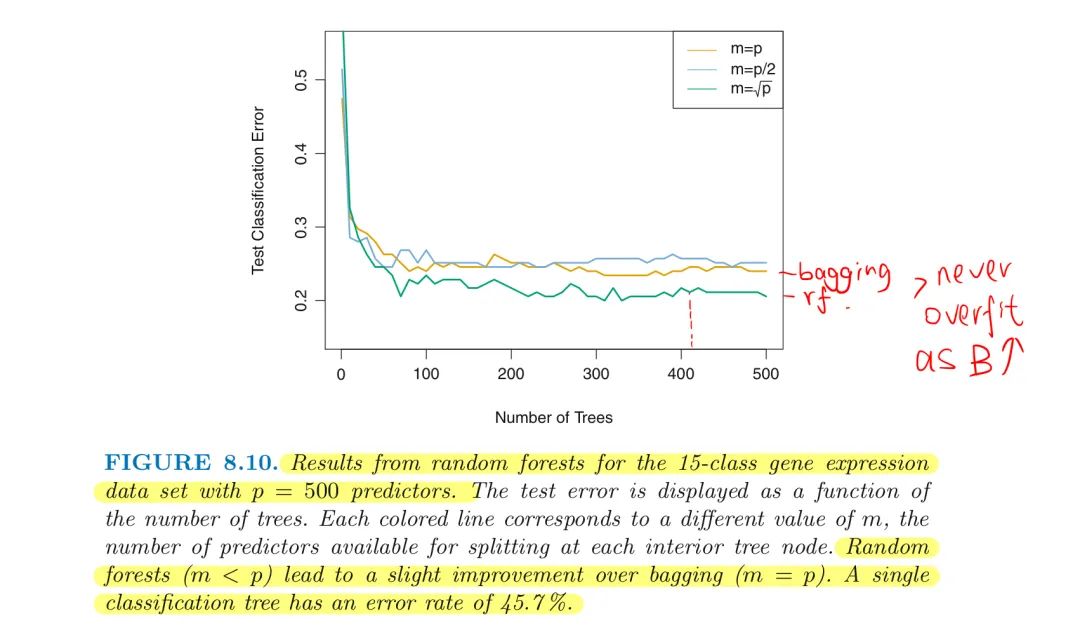
- $m\approx\sqrt{p}$
RF vs Bagging
The prediction from the bagged trees is highly correlated,
- since most trees will use the strongest predictor in the top split
- all of the bagged trees look quite similar
Decorrelating the Trees
RFs force each split to consider ONLY a subset of the predictors
- on average (p-m)/p of the splits will NOT consider the strongest predictor
- other predictors will have more of a chance to be considered at first
- this makes the average of the resulting trees less variable and more reliable
- and reduction in test error and OOB error
Small m will be helpful if a large number of correlated predictors
- Example: RF on high-dimensional biology data includes 500 genes that have the largest variance in the training set
3. Boosting
Boosting does not involve bootstrap sampling
- instead each tree is grown sequentially using information from previously grown trees
- on a modified version of the original data set
Boosting for Regression Trees:
- Set $\hat{f}(x)=0\ $ and $r_i=y_i \ $ for all i in the training set.
- For b = 1,2,…,B, repeat:
(a) Fit a tree $\hat{f}^b$ with d splits (d+1 leaves) to the training data (X,r)
(b) Update $\hat{f}\ $ by adding in a shrunken version of the new tree: $$\hat{f}(x) \leftarrow \hat{f}(x)+\lambda\hat{f}^b(x)$$ (c) Update the Residuals: $$r_i \leftarrow r_i - \lambda\hat{f}^b(x_i)$$ - Output the Boosted Model: $$\hat{f}(x)=\sum_{b=1}^B\lambda\hat{f}^b(x)$$
Explanation:
Fit a tree using the current residuals rather than Y
- then add this new tree into the fitted function to update the residuals
Splits Parameter d controls each tree to be small with a few leaves
- by fitting small trees to the residuals, we slowly improve $\hat{f}$
- in areas where it does NOT perform well
The shrinkage parameter $\lambda\ $ slows the process down even further
- allows more and different trees to attack the residuals
Idea:
The Boosting learns slowly
- Unlike fitting a single large decision tree to the data, which amounts to fitting the data hard and potentially overfitting
- Unlike in bagging, the construction of each tree depends strongly on the trees that have already been grown
- In general, Statistical Learning methods that learn slowly tend to perform well
Three Tuning Parameters:
1.The number of trees B.
-
Large B leads to overfit for boosting, unlike bagging and RFs
-
Use CV to select B
2.The shrinkage parameter $\lambda$
-
$\lambda$ = 0.01 or 0.01 typically controls the boosting learning rate
-
a very small $\lambda\ $ can require using a very large B to perform well
3.The number d of splits in each tree
-
controls the complexity of the boosted ensemble
-
$d=1$ works well, in which case each tree is stump with single split
-
the boosted ensemble is fitting an additive model, since each term involves ONLY a single variable
-
d is the interaction depth that controls the interaction order of the boosted model, since d splits can involve at most d variables
Boosting vs RF
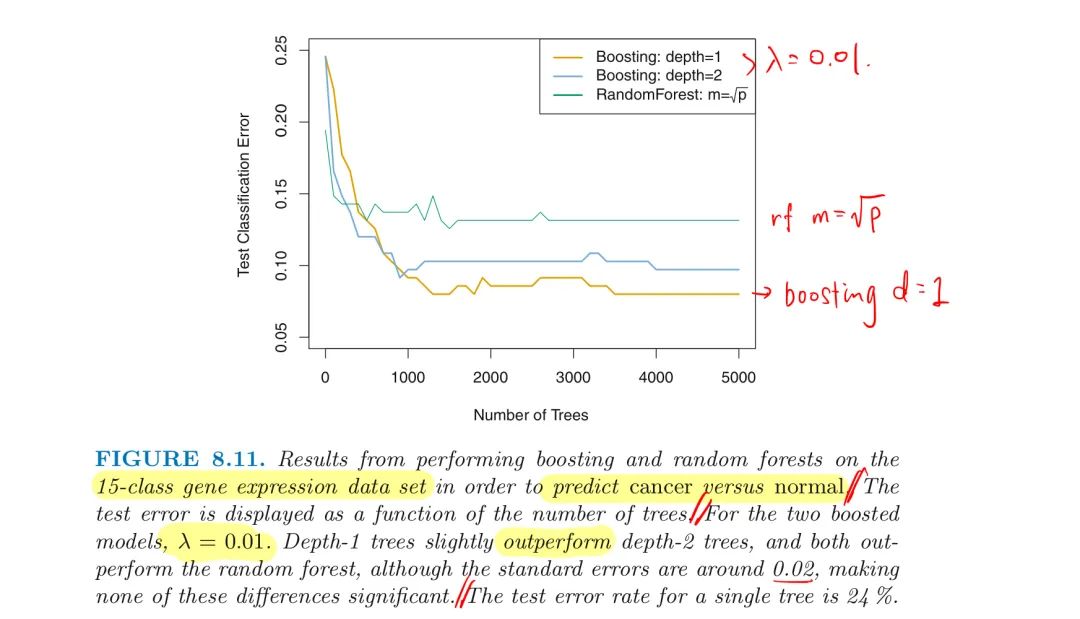
In boosting, since the growth of a particular tree takes into account the other trees that have already been grown, smaller trees are typically sufficient
- using smaller trees can aid in interpretability
- e.g. using stumps leads to an additive model