Decision Tree 1 - Regression and Classification Trees
0. Tree-based methods
Involving stratifying / segmenting the predictor space into a number of simple regions
- Use the mean/mode of the training data in the region as prediction for test data
1. Regression Decision Tree
1.1 Motivation
Making Prediction via Stratification of the Feature Space:
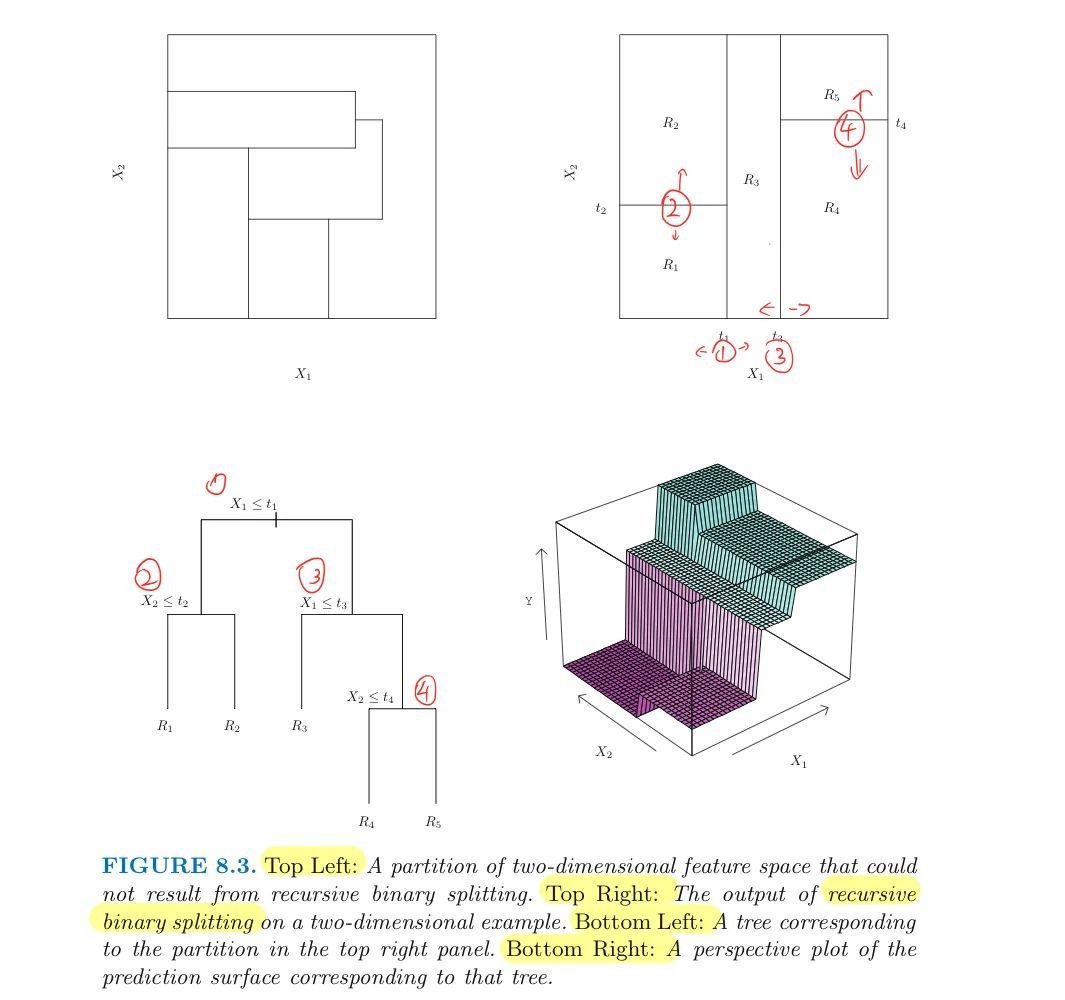
- Divide the predictor space – that is, the set of possible response $Y$ for ${X_1,X_2,…,X_p}$ – into $J$ distinct and non-overlapping regions, ${R_1,R_2,…,R_J}$
- For every test data which will fall into the region $R_j$, we make the same prediction, which is simply the mean of the response $Y$ for the training observations in $R_j$
1.2 Tree Splitting
The goal is to find leaves $R_1,…,R_J$ that minimizes the RSS, given by: $$\sum_{j=1}^J\sum_{i\in R_j}(y_i-\hat{y}_{R_j})^2$$
- where $\hat{y}_{R_j}$ is the mean response for the training observations within the $j,^{th}$ leaf
Problem: it is computationally infeasible to consider every possible partition of the feature space into $J$ leaves
Solution: Recursive Binary Splitting is a top-down, greedy approach:
- top-down because it begins at the top of the tree (all in one region) and then successively splits the predictor space;
- greedy because the best split is made at each step, rather than
looking ahead globally and picking a split will lead to a better tree in some future step(which is impossible)
First, for any feature $p$ and cutpoint $s$ , we define two regions: $$R_1(p,s)={X|X_j< s}$$ $$and$$ $$R_2(p,s)={X|X_j\geq s}$$ to get the best $p$ and $s$ that minimize: $$\sum_{i:x_i\in R_1(p,s)}(y_i-\hat{y}_{R_1})^2+\sum_{i:x_i\in R_2(p,s)}(y_i-\hat{y}_{R_2})^2$$
Next, repeat the process, looking for the best $p$ and $s$ to continue splitting
- until a stopping criterion is reached:
- e.g. no region contains more than 5 observations.
If the number of features $p$ is not too large, this process can be done quickly
- predict $Y$ in test data using the mean of the train data in the region $R_j$ to which the test data belongs
1.3 Tree Pruning
Problem:Complex Tree will lead overfit (each leaf has one data)
Solution: A smaller tree with fewer splits (fewer $R_j\ $) may lead to lower variance and better interpretation at the cost of a little bias
Method 1 - Threshold
Splitting only as the decrease in the RSS exceeds some (high) threshold
- Problem: too short-sighted since a seemingly worthless split early on may lead to a better split with large reduction in RSS
Method 2 - Pruning
Grow a very large tree $T_0\ $, then prune it back in order to obtain a subtree
- Goal: select a subtree that leads to the lowest test error rate
Rather than CVing every possible subtree, we consider a sequence of trees indexed by non-negative tuning parameter $\alpha$
- Cost Complexity / Weakest Link Pruning
For each value of $\alpha$ there corresponds a subtree $T\subset T_0$ to minimize: $$\sum_{j=1}^{|T|}\sum_{i: \ x_i\in R_j}(y_i-\hat{y}_{Rj})^2 + \alpha|T|$$
- $|T|$ : number of leaves of the tree $T$
The tuning parameter $\alpha$ controls a trade-off between the subtree’s complexity and its fit to the training data.
- $\alpha= 0 => T = T_0$, just measures the error
- As $\alpha,$ increases, there is penalty for the subtree with many leaves
- so branches get pruned from the tree in a nested and predictable fashion,
- then obtaining the whole sequence of subtrees (as a function of $\alpha$ ) is easy
$\alpha$ is similar to $\lambda$ of the lasso, which is a controller of the complexity of a linear model
- also can be selected via CV and obtain the subtree corresponding to $\alpha$
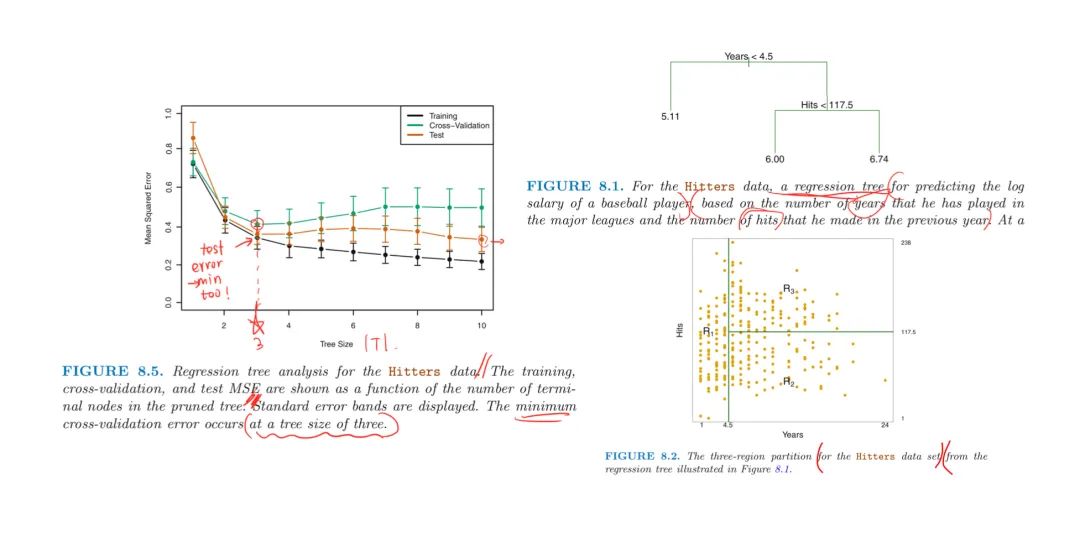
Example on Baseball Hitters Data
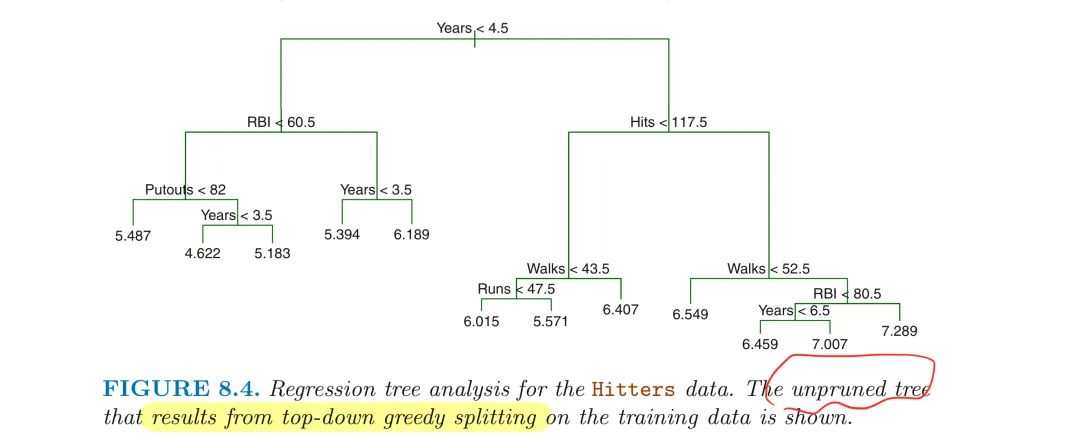
Perform 6-fold CV to estimate the CV MSE of the trees as a function of $\alpha$
- CV error is minimum at $|T|=3$ based on the best $\alpha$
2. Classification Trees
For a classification tree, we predict the test data belongs to the most commonly occurring class of train data in the region to which it belongs
- RSS cannot be a criterion for classification tree
- need two criterions to evaluate the quality of a particular split
Gini Index:
A measure of total variance across the $K$ classes:
$$G=\sum_{k=1}^K\hat{p}_{mk}(1-\hat{p}_{mk})$$
- $\hat{p}_{mk}$ represents the proportion of train data in the $m^{th}$ region that are from the $k^{th}$ class
- G small if all $\hat{p}_{mk}$ are close 1 or 0
- Node Purity: Smaller if a node contains larger amount of observations from a single class
Cross-Entropy: $$D=-\sum_{k=1}^K\hat{p}_{mk}\log (\hat{p}_{mk})$$
- Like the Gini Index, the Entropy is smaller if $m^{th}$ node is pure
- both are sensitive to node purity
Heart Disease Example:
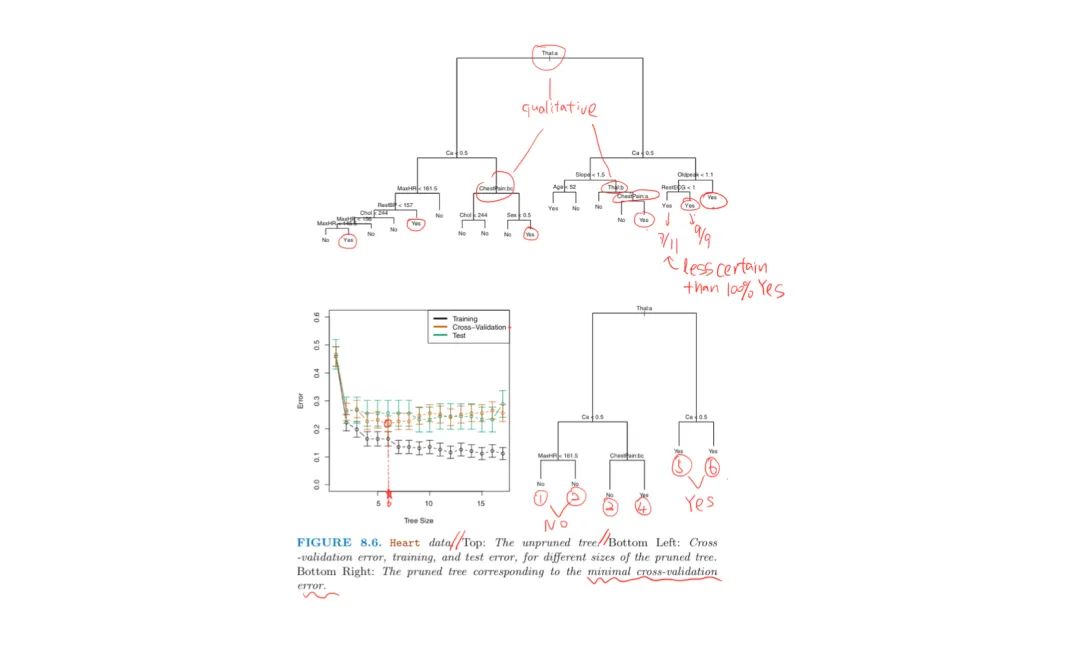
The splits may yield two same predicted value, there are reasons to keep them:
- because it leads to increased node purity
- improves the Gini Index and the Entropy
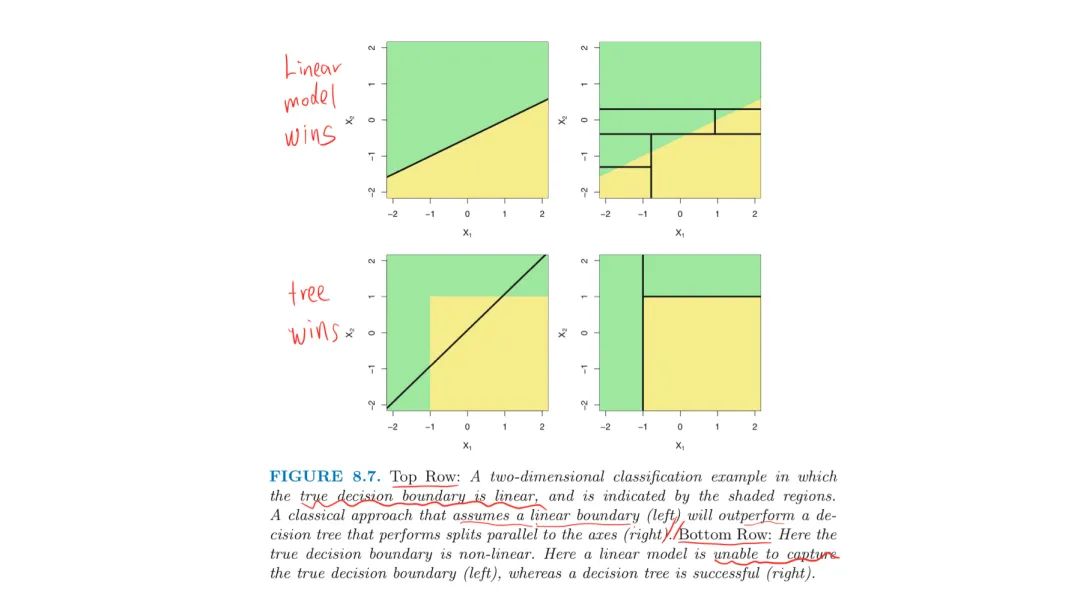
3. Trees vs Linear Models
If there is a highly non-linear complex relathinship between the features and the response, CARTs may outperform classical approaches
- However, there may still be linear relationship
4. Pros & Cons of Trees
Advantages:
- Easier to interpret than Linear Regression
- More closely mirror human decision making
- Trees can be displayed graphically
- Easily handle qualitative predictor
without the dummy variables
Limitations:
- Trees can be very non-robust:
- a small change in the data can cause a large change in the final estimated tree
- Trees do not have the same level of predictive accuracy as other regression and classification methods
- TOGO:By aggregating many decision trees, bagging, random forests and boosting will improve accuracy, at the expense of some loss in interpretation
5. Reference
An Introduction to Statistical Learning, with applications in R (Springer, 2013)